Applications of A novel alkaline metalloprotease purified by Pseudomonas aeruginosa MH298778
Main Article Content
Abstract
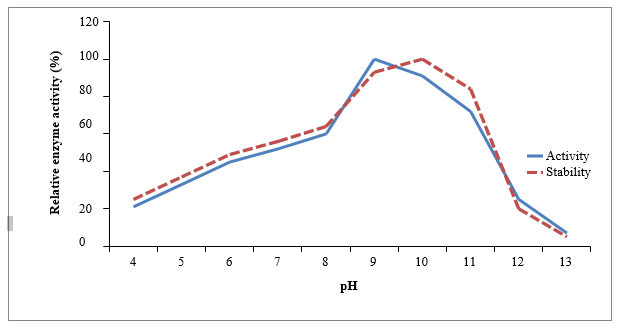
Proteases, an important class of industrially important enzymes, are widely used in detergents, leather processing, silk processing, medicine, food, animal feed, environmental protection and chemical processing. In this study an alkaline protease was isolated from the sediment sample collected from specific site of Lucknow, India and identified as Pseudomonas aeruginosa MH298778 on the basis of 16S rDNA gene sequencing and biochemical properties. The isolated protease was purified to homogeneity using ammonium sulphate precipitation, dialysis and ion exchange chromatography with a fold purification of 2.28 and a recovery of 35.91%. The enzyme has a relative molecular weight of 30 kDa, pH and temperature optima for this protease were 9.0 and 40ᵒC. The activity was stable between a pH ranged of 8.0-10.0. Metal ions such as Ca2+ and Mg2+ stimulated the protease activity up to 115 and 109% respectively. Selected metals stimulated the protease activity where as EDTA completely inhibited indicating it as 'metalloprotease'. The protease was found to be stable in some commonly used commercial detergents tested (Wheel, Rin, Ariel and Tide). It retained 70% residual activity after 3hrs of incubation with Wheel, 71% with Rin, 73% with Ariel and 52% with Tide. The maximum stability was observed with Ariel. The supplementation of the enzyme preparation in detergents could significantly improve the cleansing performance towards the blood stains and suggest its possible use as a laundry additive. The protease was also found efficient of degrading 60% of poultry feathers in 48hrs showing its keratin degrading potential therefore can also be used as an additive in poultry industry in waste treatment.
Key words: Alkaline protease, Purification, Characterization, Detergent compatibility, washing performance, feather degradation, keratin, Pseudomonas aeruginosa MH298778
Article Details
References
Abdelnasser, S.S., Ibrahim, A., Salamah, Al., Yahya, B., Elbadawi, A., Mohamed, A., Tayeb, El. and Ibrahim, S. (2015). Production of extracellular alkaline protease by new halotolerant alkaliphilic Bacillus sp. NPST-AK15 isolated from hyper saline soda lakes. Elec J of Biotechnol 18: 236–243.
Anwar, A. and Saleemuddin, M. (2000). Alkaline protease from Spilosoma obliqua: potential applications in bioformulation. Biotechnol. Appl. Biochem. 31: 85-89.
Ash, K., Sushma, Lall, A., Rao, K. and Ramteke, P. (2018). Production and optimization of an alkaline protease from Acinetobacter variabilis isolated from soil samples. International J Agri,Environ Biotechnol. 11(2): 1-8.
Ash, K., Sushma. and Ramteke, P. (2018). Optimization of extracellular alkaline protease production from Pseudomonas aeruginosa isolated from soil samples. International J Agri,Environ Biotechnol. 11(1): 187-194.
Ash, K. and Mishra, S.K. (2023). Protease Enzymes: Present Status and Future Perspectives for Industrial Sector. Int. J. Curr. Microbiol. App. Sci, 12(2): 311-323.
Banerjee, U.C., Sani, R.K., Azmi, W. and Soni, R. (1999). Thermostable alkaline protease from Bacillus brevis and its characterization as a laundry detergent additive. Proces Biochem. 35:213-219.
Baweja, M., Singh, P., Sadaf, A., Tiwari, R., Nain, L., Khare, S. and Shukla, P. (2017). Cost effective characterization process and molecular dynamic simulation of detergent compatible alkaline protease from Bacillus pumilus strain MP27. Process Biochem. 58: 199-203.
Bayoudh, A., Manni, L., Agrebi, R. and Nasri, M. (2008). Purification, biochemical and molecular characterization of a metalloprotease from Pseudomonas aeruginosa MN7 grown on shrimp wastes. Applied Microbiol and Biotechnol. 79(6):989-995.
Drapeau, G., Boily, Y. and Houmard, J. (1972). Purification and properties of an extracellular protease of Staphylococcus aureus. J Biological chem. 247(20):6720-6726.
Elibol, M. and Moreira, A.R. (2005). Optimizing some factors affecting alkaline protease production by a marine bacterium Teredinobacter turnirae under solid substrate fermentation. Pro Biochem. 40: 1-6.
Fritz, J.S. (2004). Early milestones in the development of ion-exchange chromatography: a personal account. J Chromatography. 1039 (1-2):3-12.
Furhan, J. and Sharma, S. (2014). Microbial alkaline proteases: findings and applications.
Int. J. Inv. Pharm. Sci. 2(4):823-834.
Ghafoor, A. and Hasnain, S. (2010). Purification and characterization of an extracellular protease from Bacillus subtilis EAG-2 strain isolated from ornamental plant nursery. Pol J Microbiol. 59(2):107-112.
Giri, S.B., Sukumaran, V., Sen, S.S., Oviya, M., Banu, B.N. and Jena, P.K. (2011). Purification and partial characterization of a detergent and oxidizing agent stable alkaline protease from a newly isolated Bacillus subtilis VSG-4 of tropical soil. The J Microbiol. 49(3): 455-461.
Gupta, M., Aggarwal, S., Navani, N. and Choudhury, B. (2015). Isolation and characterization of a protease-producing novel haloalkaliphilic bacterium Halobiforma sp. strain BNMIITR from Sambhar lake in Rajasthan, India. Annals of Microbiol. 65(2): 677-686.
Gupta, R., Beg, Q.K. and Lorenz, P. (2002). Bacterial alkaline proteases: molecular approaches and industrial applications. Appl Microbiol Biotechnol. 59: 15-32.
Haddar, A., Kamoun, A.S., Zouari, N.F., Hmidet, N. and Nasri, M. (2010). Characterization of detergent stable and feather degrading serine proteases from Bacillus mojavensis A 21. Biochem Eng J.51:53-63.
Hamamoto, T., Kaneda, M., Horikoshi, K. and Kudo, T. (1994). Characterization of a protease from a psychrotroph Pseudomonas fluorescens 114. Appl. Environ. Microbiol. 60: 3878-3880.
Jisha, N.V., Smitha, R.B., Pradeep, S., Sreedevi, S., Unni, K.N., Sajith, S., Priji, P., Josh,
M.S. and Benjamin, S. (2013). Versatility of microbial proteases. Advan. in Enzymes Research. 1(3): 39-51.
Krishna, K.V., Gupta, M., Gupta, N., Gaudani, H., Trivedi, S., Patil, P., Gupta, G., Khairnar, Y., Borasate, A. and Mishra, D. (2009). Alkaline proteases. International J Microbiol Res. 1: 14-18.
Kuddus, M. and Ramteke, P. W. (2008). A cold active- extracellular metalloprotease from Curtobacterium luteum (MTCC 7529): enzyme production and characterization. J. Gen. Appl. Microbiol. 54: 393-398.
Kuddus, M. and Ramteke, P. W. (2009). A cold active- extracellular alkaline protease from an alkaliphilic Stenotrophomonas maltophilia: production of enzyme and its industrial applicatios . Can. J. Microbiol. 55:1-8.
Kuddus, M. and Ramteke, P. W. (2011). Production optimization of an extracellular cold active alkaline protease from Stenotrophomonas maltophilia MTCC 7528 and its application in detergent industry. African J of Microbiol. 5(7): 809-816.
Kuddus, M., Joseph, B. and Ramteke, P. W. (2013). Production of laccase from newly isolated Pseudomonas putida and its application in bioremediation of synthetic dyes and industrial effluents. Biocatalysis and Agriculture Biotechnol. 2: 333-338.
Kumar, R. and Bhalla, T.C. (2004). Purification and characterization of a small size protease from Bacillus sp. Indian J Exp Biol. 42(5): 15-21.
Kumari, B., Singh, S.N. and Singh, D.P. (2012). Characterization of two biosurfactant producing strains in crude oil degradation. Proc Bioche .47(12): 2463-2471.
Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227(5259): 680-685.
Li, G.Y., Cai, Y.J., Liao, X.R. and Yin, J. (2011). A novel nonionic surfactant- and solvent-stable alkaline serine protease from Serratia sp. SYBC H with duckweed as nitrogen source: production, purification, characteristics and application. J Ind.Microbiol. Biotechnol.38:845-853.
Lowry, O.H., Rosebrough, N.J., Farr, A.L., Randal, R.J. (1951). Protein measurement with the folin phenol reagent. J.Biol.Chem. 193:265-275.
Naby, M., Ahmed, S., Wehaidy, H. and Mahdy, S. (2017). Catalytic, kinetic and thermodynamic properties of stabilized Bacillus stearothermophilus alkaline protease. International J Biol Macromolecules. 96: 265-271.
Najafi, M.F., Deobagkar, D. and Deobagkar, D. (2005). Potential application of protease isolated from Pseudomonas aeruginosa PD 100. Electr J Biotechnol. 8(2):197-203.
Nam, G.W., Lee, D.W., Lee, H.S., Kim, B.C., Choe, E.A., Hwang, J.K., Suhartono, M.T. and Pyun, Y.R. (2002). Native-feather degradation by Fervidobacterium islandicum AW- 1, a newly isolated keratinase-producing thermophilic anaerobe. Arch Microbiol.178:538- 547.
Pathak, A.P. and Deshmukh, K.B. (2012). Alkaline protease production, extraction and characterization from alkaliphilic Bacillus licheniformis KBDL4: A lonar soda lake isolate. Indian J Experimental Biol. 50: 569-576.
Patil, R.C. and Jadhav, B.L. (2017). Isolation and Characterization of Protease Producing Bacillus Species from Soil of Dairy Industry. Int.J.Curr.Microbiol.App.Sci., 6(6): 853- 860.
Priji, P., Sajith, S., Faisal, P. and Benjamin, S. (2017). Psedomonas sp. BUP6 produces a thermotolerant alkaline lipase with transesterification efficiency in producing biodiesel. 3Biotech. 369(7): 21-30.
Rai, S.K. and Mukherjee, A.K. (2010). Statistical optimization of production, purification and industrial application of a laundry detergent and organic solvent-stable subtilisin-like serine protease (alzwiprase) from Bacillus subtilis DM-04. Biochem Eng J.48:173-180.
Rawlings, N.D., Barrett, A.J. and Bateman, A. (2011). Asparagine peptidase lyases: a seventh catalytic type of proteolytic enzymes. J Biol Chem 286:38321-38328.
Romero, I., Ignacio, J., Fernandez, F. and Garcia, J. (2008). Characterization of the main enzymatic activities present in Serratia marcescens and their effects on destaining. International J Sci Technol. 43(7):1295-1305.
Sana, B., Ghosh, D., Saha, M. and Mukherjee, J. (2006). Purification and characterization of a salt, solvent, detergent and bleach tolerant protease from a new gamma- Proteobacterium isolated from the marine environment of the Sunderbans. Proc Biochem.41: 208-215.
Sawant, R. and Nagendran, S. (2014). Protease: An Enzyme with multiple industrial applications. World J of Pharmacy and Pharmaceutical Sci. 3(6): 568-579.
Secades, P. and Guijarro, J.A. (1999). Purification and characterization of an extracellular protease from the fish pathogen Yersinia ruckeri and effect of culture conditions on production. Appl. Environ. Microbiol. 65(9):3969-3975.
Sharma, K.M., Kumar, R., Panwar, S. and Kumar, A. (2017). Microbial alkaline protases: optimization of production parameters and their properties. J Genetic Engg.Biotechnol.15:115-126.
Shine, K., Kanimozhi, K., Panneerselvam, A., Muthukumar, C. and Thajuddin, N. (2016). Production and optimization of alkaline protease by Bacillus cereus RS3 isolated from desert soil. Int. J. Adv. Res. Biol. Sci. 3(7): 193-202.
Sinha, P., Singh, R.K., Srivastava, R., Sharma, R. and Prakash, S. (2013). Characterization and optimization of alkaline protease enzyme produced by soil borne bacteria. Trends Life Sci International J. 2(2):219-224.
Tamilmani, P., Umamaheswari, A., Vinayagam, A. and Prakash, B. (2008). Production of an extracellular feather degrading enzyme by Bacillus licheniformis isolated from poultry farm soil in Namakkal District (Tamilnadu). International J Poultry Science. 7(2):184- 188.
Xin, Xiong., Ambati, Rao., Cai, Zongwei. and Lei, Bo. (2018). Purification and characterization of fibrinolytic enzyme from a bacterium isolated from soil. 3 Biotech. 90(8): 219-225.
Xu, J., Jiang, M., Honglin, S. and He, B. (2010). An organic solvent-stable protease from organic solvent- tolerant Bacillus cereus WQ9-2: purification, biochemical properties, and potential application in peptide synthesis. Bioresource Technol. 101: 7991-7994.
Zambare, V., Nilegaonkar, S. and Anovel, P.K. (2011). New Biotechnol. http://dx.
doi.org/10.1016/j.nbt.2010.10.002.